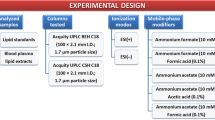
Abstract
Reversed-phase ultrahigh-performance liquid chromatography-mass spectrometry (RP-UHPLC/MS) method was developed with the aim to unambiguously identify a large number of lipid species from multiple lipid classes in human plasma. The optimized RP-UHPLC/MS method employed the C18 column with sub-2-μm particles with the total run time of 25 min. The chromatographic resolution was investigated with 42 standards from 18 lipid classes. The UHPLC system was coupled to high-resolution quadrupole-time-of-flight (QTOF) mass analyzer using electrospray ionization (ESI) measuring full-scan and tandem mass spectra (MS/MS) in positive- and negative-ion modes with high mass accuracy. Our identification approach was based on m/z values measured with mass accuracy within 5 ppm tolerance in the full-scan mode, characteristic fragment ions in MS/MS, and regularity in chromatographic retention dependences for individual lipid species, which provides the highest level of confidence for reported identifications of lipid species including regioisomeric and other isobaric forms. The graphs of dependences of retention times on the carbon number or on the number of double bond(s) in fatty acyl chains were constructed to support the identification of lipid species in homologous lipid series. Our list of identified lipid species is also compared with previous publications investigating human blood samples by various MS-based approaches. In total, we have reported more than 500 lipid species representing 26 polar and nonpolar lipid classes detected in NIST Standard reference material 1950 human plasma.
Graphical abstract








Similar content being viewed by others
Abbreviations
- BPI:
-
Base peak intensity
- CAR:
-
Acylcarnitine
- CE:
-
Cholesteryl ester
- Cer:
-
Ceramide
- CN:
-
Carbon number
- DB:
-
Double bond
- DG:
-
Diacylglycerol
- DI:
-
Direct infusion
- ECN:
-
Equivalent carbon number
- ESI:
-
Electrospray ionization
- FA:
-
Fatty acid
- GlcCer:
-
Glucosylceramide
- GM3:
-
Monosialodihexosylganglioside
- HexCer:
-
Hexosylceramide
- Hex2Cer:
-
Dihexosylceramide
- Hex3Cer:
-
Trihexosylceramide
- Hex4Cer:
-
Tetrahexosylceramide
- HILIC:
-
Hydrophilic interaction liquid chromatography
- HPLC:
-
High-performance liquid chromatography
- Chol:
-
Cholesterol
- IS:
-
Internal standard
- LacCer:
-
Lactosylceramide
- LPC:
-
Lysophosphatidylcholine
- LPE:
-
Lysophosphatidylethanolamine
- LPG:
-
Lysophosphatidylglycerol
- LPI:
-
Lysophosphatidylinositol
- LPS:
-
Lysophosphatidylserine
- MG:
-
Monoacylglycerol
- MS:
-
Mass spectrometry
- MS/MS:
-
Tandem mass spectrometry
- NP:
-
Normal-phase
- -O:
-
Alkyl bond
- -P:
-
Plasmalogen
- PC:
-
Phosphatidylcholine
- PE:
-
Phosphatidylethanolamine
- PG:
-
Phosphatidylglycerol
- PI:
-
Phosphatidylinositol
- PS:
-
Phosphatidylserine
- RIC:
-
Reconstructed ion current
- RP:
-
Reversed-phase
- SM:
-
Sphingomyelin
- TG:
-
Triacylglycerol
- tR :
-
Retention time
- UHPLC:
-
Ultrahigh-performance liquid chromatography
- UHPSFC:
-
Ultrahigh-performance supercritical fluid chromatography
- QTOF:
-
Quadrupole-time-of-flight
References
Wenk MR. The emerging field of lipidomics. Nat Rev Drug Discov. 2005;4:594–610.
Arneth B, Arneth R, Shams M. Metabolomics of type 1 and type 2 diabetes. Int J Mol Sci. 2019;20:2467.
Butler LM, Perone Y, Dehairs J, Lupien LE, de Laat V, Talebi A, et al. Lipids and cancer: emerging roles in pathogenesis, diagnosis and therapeutic intervention. Adv Drug Deliv Rev. 2020;159:245–93.
Huynh K, Barlow CK, Jayawardana KS, Weir JM, Mellett NA, Cinel M, et al. High-throughput plasma lipidomics: detailed mapping of the associations with cardiometabolic risk factors. Cell Chem Biol. 2019;26:71–84.
Laaksonen R, Ekroos K, Sysi-Aho M, Hilvo M, Vihervaara T, Kauhanen D, et al. Plasma ceramides predict cardiovascular death in patients with stable coronary artery disease and acute coronary syndromes beyond LDL-cholesterol. Eur Heart J. 2016;37:1967–76.
Tonks KT, Coster AC, Christopher MJ, Chaudhuri R, Xu A, Gagnon-Bartsch J, et al. Skeletal muscle and plasma lipidomic signatures of insulin resistance and overweight/obesity in humans. Obesity. 2016;24:908–16.
van Kruining D, Luo Q, van Echten-Deckert G, Mielke MM, Bowman A, Ellis S, et al. Sphingolipids as prognostic biomarkers of neurodegeneration, neuroinflammation, and psychiatric diseases and their emerging role in lipidomic investigation methods. Adv Drug Deliv Rev. 2020;159:232–44.
Wolrab D, Jirásko R, Chocholoušková M, Peterka O, Holčapek M. Oncolipidomics: mass spectrometric quantitation of lipids in cancer research. TrAC - Trends Anal Chem. 2019;120:115480.
Wang J, Wang C, Han X. Tutorial on lipidomics. Anal Chim Acta. 2019;1061:28–41.
Wolrab D, Jirásko R, Cífková E, Höring M, Mei D, Peterka O, et al. Lipidomic profiling of human serum enables detection of pancreatic cancer 1. Nat Com. 2021; revision; preprint available at https://www.medrxiv.org/content/10.1101/2021.01.22.21249767v1
LIPID MAPS. LIPID MAPS Lipidomics Gateway. 2021 (accessed 5.5.2021). https://lipidmaps.org/
Holčapek M, Liebisch G, Ekroos K. Lipidomic analysis. Anal Chem. 2018;90:4249–57.
Hsu FF. Mass spectrometry-based shotgun lipidomics – a critical review from the technical point of view. Anal Bioanal Chem. 2018;410:6387–409.
Sales S, Graessler J, Ciucci S, Al-Atrib R, Vihervaara T, Schuhmann K, et al. Gender, contraceptives and individual metabolic predisposition shape a healthy plasma lipidome. Sci Rep. 2016;6:27710.
Han X, Yang K, Gross RW. Multi-dimensional mass spectrometry-based shotgun lipidomics and novel strategies for lipidomic analyses. Mass Spectrom Rev. 2012;31:134–78.
Čajka T, Fiehn O. Comprehensive analysis of lipids in biological systems by liquid chromatography-mass spectrometry. TrAC – Trends Anal Chem. 2014;61:192–206.
Triebl A, Hartler J, Trötzmüller MC, Köfeler H. Lipidomics: prospects from a technological perspective. Biochim Biophys Acta Mol Cell Biol Lipids. 1862;2017:740–6.
Cífková E, Holčapek M, Lísa M, Vrána D, Melichar B, Študent V. Lipidomic differentiation between human kidney tumors and surrounding normal tissues using HILIC-HPLC/ESI-MS and multivariate data analysis. J Chromatogr B. 2015;1000:14–21.
Wolrab D, Chocholoušková M, Jirásko R, Peterka O, Holčapek M. Validation of lipidomic analysis of human plasma and serum by supercritical fluid chromatography–mass spectrometry and hydrophilic interaction liquid chromatography–mass spectrometry. Anal Bioanal Chem. 2020;412:75–2388.
Rampler E, Schoeny H, Mitic BM, el Abiead Y, Schwaiger M, Koellensperger G. Simultaneous non-polar and polar lipid analysis by on-line combination of HILIC, RP and high resolution MS. Analyst. 2018;143:1250–8.
Dispas A, Jambo H, André S, Tyteca E, Hubert P. Supercritical fluid chromatography: a promising alternative to current bioanalytical techniques. Bioanalysis. 2018;10:107–24.
Lísa M, Holčapek M. High-throughput and comprehensive lipidomic analysis using ultrahigh-performance supercritical fluid chromatography-mass spectrometry. Anal Chem. 2015;87:7187–95.
Chocholoušková M, Jirásko R, Vrána D, Gatěk J, Melichar B, Holčapek M. Reversed phase UHPLC/ESI-MS determination of oxylipins in human plasma: a case study of female breast cancer. Anal Bioanal Chem. 2019;411:1239–51.
Ovčačíková M, Lísa M, Cífková E, Holčapek M. Retention behavior of lipids in reversed-phase ultrahigh-performance liquid chromatography-electrospray ionization mass spectrometry. J Chromatogr A. 2016;1450:76–85.
Damen CWN, Isaac G, Langridge J, Hankemeier T, Vreeken RJ. Enhanced lipid isomer separation in human plasma using reversed-phase UPLC with ion-mobility/high-resolution MS detection. J Lipid Res. 2014;55:1772–83.
Yamada T, Uchikata T, Sakamoto S, Yokoi Y, Fukusaki E, Bamba T. Development of a lipid profiling system using reverse-phase liquid chromatography coupled to high-resolution mass spectrometry with rapid polarity switching and an automated lipid identification software. J Chromatogr A. 2013;1292:211–8.
Fauland A, Köfeler H, Trötzmüller M, Knopf A, Hartler J, Eberl A, et al. A comprehensive method for lipid profiling by liquid chromatography-ion cyclotron resonance mass spectrometry. J Lipid Res. 2011;52:2314–22.
Sandra K, Pereira AS, Vanhoenacker G, David F, Sandra P. Comprehensive blood plasma lipidomics by liquid chromatography/quadrupole time-of-flight mass spectrometry. J Chromatogr A. 1217:4087–99.
Lísa M, Holčapek M. Triacylglycerols profiling in plant oils important in food industry, dietetics and cosmetics using high-performance liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. J Chromatogr A. 2008;1198–1199:115–30.
Nikolova-Damyanova B. Retention of lipids in silver ion high-performance liquid chromatography: facts and assumptions. J Chromatogr A. 2009;1216:1815–24.
Lísa M, Denev R. Holčapek M (2013) Retention behavior of isomeric triacylglycerols in silver-ion HPLC: effects of mobile phase composition and temperature. J Sep Sci. 2013;36:2888–900.
Seon HL, Williams M v, Blair IA. Targeted chiral lipidomics analysis. Prostaglandins Other Lipid Mediat. 2005;77:141–57.
Lísa M, Holčapek M. Characterization of triacylglycerol enantiomers using chiral HPLC/APCI-MS and synthesis of enantiomeric triacylglycerols. Anal Chem. 2013;85:1852–9.
Holčapek M, Jandera P, Fischer J, Prokeš B. Analytical monitoring of the production of biodiesel by high-performance liquid chromatography with various detection methods. J Chromatogr A. 1999;858:13–31.
Holčapek M, Ovčačíková M, Lísa M, Cífková E, Hájek T. Continuous comprehensive two-dimensional liquid chromatography–electrospray ionization mass spectrometry of complex lipidomic samples. Anal Bioanal Chem. 2015;407:5033–43.
Murphy RC Tandem mass spectrometry of lipids: molecular analysis of complex lipids. 1st ed. Cambridge; Royal Society of Chemistry; 2014.
Lísa M, Cífková E, Holčapek M. Lipidomic profiling of biological tissues using off-line two-dimensional high-performance liquid chromatography-mass spectrometry. J Chromatogr A. 2011;1218:5146–56.
Hořejší K, Jirásko R, Chocholoušková M, Wolrab D, Kahoun D, Holčapek M. Comprehensive identification of glycosphingolipids in human plasma using hydrophilic interaction liquid chromatography-electrospray ionization mass spectrometry. Metabolites. 2021;11:140–63.
Merrill AH, Sullards MC, Allegood JC, Kelly S, Wang E. Sphingolipidomics: High-throughput, structure-specific, and quantitative analysis of sphingolipids by liquid chromatography tandem mass spectrometry. Methods. 2005;36:207–24.
Liebisch G, Fahy E, Aoki J, Dennis EA, Durand T, Ejsing CS, et al. Update on LIPID MAPS classification, nomenclature, and shorthand notation for MS-derived lipid structures. J Lipid Res. 2020;61:1539–55.
Quehenberger O, Armando AM, Brown AH, Milne SB, Myers DS, Merrill AH, et al. Lipidomics reveals a remarkable diversity of lipids in human plasma1. J Lipid Res. 2010;51:3299–305.
Bowden JA, Heckert A, Ulmer CZ, Jones CM, Koelmel JP, Abdullah L, et al. Harmonizing lipidomics: NIST interlaboratory comparison exercise for lipidomics using SRM 1950-Metabolites in frozen human plasma. J Lipid Res. 2017;58:2275–88.
Wolrab D, Chocholoušková M, Jirásko R, Peterka O, Mužáková V, Študentová H, et al. Determination of one year stability of lipid plasma profile and comparison of blood collection tubes using UHPSFC/MS and HILIC-UHPLC/MS. Anal Chim Acta. 2020;1137:74–84.
Liebisch G, Ahrends R, Arita M, Arita M, Bowden JA, Ejsing CS, et al. Lipidomics needs more standardization. Nat Metab. 2019;1:745–7.
Liebisch G, Ejsing CS, Ekroos K. Identification and annotation of lipid species in metabolomics studies need improvement. Clin Chem. 2015;61:1542–4.
Kofeler HC, Eichmann TO, Ahrends R, Bowden JA, Danne-Rasche N, Fedorova M, et al. Nat Com. 2021; revision; preprint available at https://zenodo.org/record/4672232#.YJ5c0odxeUl
Funding
This work was supported by the grant project no. 21-20238S funded by the Czech Science Foundation.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Ethics approval
The ethical approval is not needed for this study, because biological samples of patients or clinical data are not used in this study.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Published in the topical collection celebrating ABCs 20th Anniversary.
Rights and permissions
About this article
Cite this article
Vaňková, Z., Peterka, O., Chocholoušková, M. et al. Retention dependences support highly confident identification of lipid species in human plasma by reversed-phase UHPLC/MS. Anal Bioanal Chem 414, 319–331 (2022). https://doi.org/10.1007/s00216-021-03492-4
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00216-021-03492-4